Volume 9, Issue 3 (Journal of Clinical and Basic Research (JCBR) 2025)
jcbr 2025, 9(3): 1-5 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Rakhshi R, Khosravi T, Rahimzadeh A, Mohsenipour M, Oladnabi M. DeepSeek-R1: Reliability for research and education – A comparative study with Claude-3.5-Sonnet and GPT-4o. jcbr 2025; 9 (3) :1-5
URL: http://jcbr.goums.ac.ir/article-1-516-en.html
URL: http://jcbr.goums.ac.ir/article-1-516-en.html
Reza Rakhshi1 

 , Teymoor Khosravi1
, Teymoor Khosravi1 

 , Arian Rahimzadeh1
, Arian Rahimzadeh1 

 , Mohadeseh Mohsenipour1
, Mohadeseh Mohsenipour1 

 , Morteza Oladnabi *2
, Morteza Oladnabi *2 




 , Teymoor Khosravi1
, Teymoor Khosravi1 

 , Arian Rahimzadeh1
, Arian Rahimzadeh1 

 , Mohadeseh Mohsenipour1
, Mohadeseh Mohsenipour1 

 , Morteza Oladnabi *2
, Morteza Oladnabi *2 


1- Student Research Committee, Golestan University of Medical Sciences, Gorgan, Iran
2- Gorgan Congenital Malformations Research Center, Jorjani Clinical sciences Research Institute, Golestan University of Medical Sciences, Gorgan, Iran; Department of Medical Genetics, School of Advanced Technologies in Medicine, Golestan University of Medical Sciences, Gorgan, Iran ,oladnabidozin@yahoo.com
2- Gorgan Congenital Malformations Research Center, Jorjani Clinical sciences Research Institute, Golestan University of Medical Sciences, Gorgan, Iran; Department of Medical Genetics, School of Advanced Technologies in Medicine, Golestan University of Medical Sciences, Gorgan, Iran ,
Full-Text [PDF 391 kb]
(821 Downloads)
| Abstract (HTML) (1154 Views)
Discussion
DeepSeek-R1 emerges as a robust alternative to proprietary LLMs, particularly in resource-constrained academic environments. Its MoE architecture and multi-stage training (Cold-start SFT + RL) enable efficient reasoning, as evidenced by benchmark outperformance in math and coding (3,5). These results align with recent studies on instruction-tuned models achieving 85-89% USMLE accuracy (4,6,9), underscoring its utility in medical education and research.
The user study corroborates these findings, with comparable diagnostic accuracy across specialties, suggesting DeepSeek-R1's viability for clinical simulations and knowledge dissemination. However, its conciseness may limit narrative tasks, contrasting with more verbose models like GPT-4o (5). Cost savings (96%) enhance accessibility, but censorship poses risks for unbiased research in social sciences (8,11).
Data privacy concerns, due to Chinese servers, warrant caution for sensitive data (12). Compared to Claude-3.5-Sonnet's detailed bias mitigation, DeepSeek-R1's documentation is sparse, potentially amplifying ethical issues (10). Future work should explore fine-tuning to mitigate censorship and extend output limits. Overall, DeepSeek-R1's open-source nature democratizes AI (7), but users must cross-verify outputs and consider hybrid approaches for comprehensive applications.
Limitations
However, concerns have been raised regarding its reliability for research and educational purposes, particularly due to its implementation of censorship mechanisms that restrict responses on politically sensitive topics, as well as potential data privacy and security issues stemming from data storage on servers located in China.
Conclusion
DeepSeek-R1 offers reliable performance for research and education, excelling in mathematical reasoning (97.3% MATH-500), programming (96.3% HumanEval), and medical diagnostics (79.2% accuracy in user study). Its open-source framework and substantial cost efficiency (96% cheaper than GPT-4o) position it as a compelling alternative to Claude-3.5-Sonnet and GPT-4o, particularly in academic and clinical settings. However, limitations such as censorship on sensitive topics, data storage risks in China, and output restrictions (4k tokens) necessitate careful consideration. The comparative analyses (Tables 1-7, Figures 1-2) provide a framework for context-specific adoption, emphasizing the need for ethical oversight and verification in AI-assisted workflows. Ultimately, DeepSeek-R1 advances open-source AI, fostering global collaboration while highlighting the balance between innovation and security.
Acknowledgement
We are grateful to the Medical Genetics Department of Golestan University of Medical Sciences for their support and assistance.
Funding sources
This work was supported by the Golestan University of Medical Sciences and Health Services (Grant number: 113845)
Ethical statement
This study was ethically approved by Ethics Committee of Golestan University of Medical Sciences (Ethics Code: IR.GOUMS.REC.1402.458)
Conflicts of interest
No potential conflict of interest was reported by the author(s).
Author contributions
Conceptualization: Reza Rakhshi, Teymoor Khosravi; Data Curation: Teymoor Khosravi, Mohadeseh Mohsenipour, Arian Rahimzadeh; Formal Analysis: Arian Rahimzadeh, Mohadeseh Mohsenipour; Funding Acquisition: Morteza Oladnabi, Teymoor Khosravi; Investigation: Arian Rahimzadeh; Methodology: Reza Rakhshi; Project Administration: Morteza Oladnabi; Resources: Teymoor Khosravi; Software: Teymoor Khosravi, Arian Rahimzadeh; Supervision: Morteza Oladnabi; Validation: Reza Rakhshi, Mohadeseh Mohsenipour; Visualization: Reza Rakhshi, Teymoor Khosravi Arian Rahimzadeh, Mohadeseh Mohsenipour; Writing - Original Draft: Reza Rakhshi, Teymoor Khosravi, Arian Rahimzadeh; Writing - Review & Editing: Reza Rakhshi, Teymoor Khosravi, Morteza Oladnabi.
Data availability statement
No additional data were created or used in this study beyond what is presented in the manuscript.
Full-Text: (271 Views)
Introduction
In recent years, artificial intelligence (AI) has increasingly permeated various sectors, including healthcare, education, and research. AI-powered large language models (LLMs), often referred to as chatbots, have emerged as transformative tools for disseminating information, aiding in data analysis, and supporting decision-making processes (1,2). These models leverage vast datasets and advanced algorithms to generate human-like responses, enabling applications in complex tasks such as mathematical problem-solving, programming, and medical diagnostics.
A notable entrant in this domain is DeepSeek-R1, developed by DeepSeek-AI, a Chinese tech startup. DeepSeek-R1 is designed to enhance reasoning capabilities through a unique multi-stage training pipeline that integrates reinforcement learning (RL) and supervised fine-tuning (SFT), while promoting open-source collaboration (3). Unlike proprietary models like Claude-3.5-Sonnet (Developed by Anthropic) and GPT-4o (Developed by OpenAI), DeepSeek-R1 is fully open-source under the MIT license, making it accessible for modification and deployment in resource-limited settings.
The rapid evolution of LLMs has sparked interest in their reliability for academic and research purposes. For instance, models like GPT-4o have demonstrated strong performance in benchmarks such as MMLU (Massive Multitask Language Understanding) and MedQA (Medical Question Answering), but they come with high computational costs and proprietary restrictions (4,5). Claude-3.5-Sonnet offers balanced reasoning with ethical safeguards, yet it lacks the transparency of open-source alternatives (6). DeepSeek-R1 addresses these gaps by achieving comparable or superior results in structured tasks, such as mathematical reasoning (97.3% on MATH-500) and programming (96.3% on HumanEval), at a fraction of the cost (3).
However, the adoption of LLMs in research and education is not without challenges. Issues such as data privacy, censorship, and output limitations can impact their utility, particularly in sensitive fields like medicine and social sciences (7,8). This study aimed to evaluate DeepSeek-R1's reliability for research and education by comparing it with Claude-3.5-Sonnet and GPT-4o across technical benchmarks, user studies, and practical considerations. We explored its strengths in efficiency and performance, while addressing potential limitations like censorship and data security. Through this comparative analysis, we provided evidence-based insights to guide academics, researchers, and educators in adopting DeepSeek-R1, especially in medical and scientific contexts.
Methods
This study employed a mixed-methods approach, combining benchmark evaluations, a prospective user study, and comparative analyses to assess DeepSeek-R1's reliability relative to Claude-3.5-Sonnet and GPT-4o. The methodology was structured to ensure reproducibility and alignment with ethical standards.
Model selection and benchmarking
Three frontier LLMs were selected: DeepSeek-R1 (236B-parameter Mixture-of-Experts architecture, open-source under MIT license), Claude-3.5-Sonnet (Proprietary, Anthropic), and GPT-4o (Proprietary, OpenAI). Benchmarks included:
In recent years, artificial intelligence (AI) has increasingly permeated various sectors, including healthcare, education, and research. AI-powered large language models (LLMs), often referred to as chatbots, have emerged as transformative tools for disseminating information, aiding in data analysis, and supporting decision-making processes (1,2). These models leverage vast datasets and advanced algorithms to generate human-like responses, enabling applications in complex tasks such as mathematical problem-solving, programming, and medical diagnostics.
A notable entrant in this domain is DeepSeek-R1, developed by DeepSeek-AI, a Chinese tech startup. DeepSeek-R1 is designed to enhance reasoning capabilities through a unique multi-stage training pipeline that integrates reinforcement learning (RL) and supervised fine-tuning (SFT), while promoting open-source collaboration (3). Unlike proprietary models like Claude-3.5-Sonnet (Developed by Anthropic) and GPT-4o (Developed by OpenAI), DeepSeek-R1 is fully open-source under the MIT license, making it accessible for modification and deployment in resource-limited settings.
The rapid evolution of LLMs has sparked interest in their reliability for academic and research purposes. For instance, models like GPT-4o have demonstrated strong performance in benchmarks such as MMLU (Massive Multitask Language Understanding) and MedQA (Medical Question Answering), but they come with high computational costs and proprietary restrictions (4,5). Claude-3.5-Sonnet offers balanced reasoning with ethical safeguards, yet it lacks the transparency of open-source alternatives (6). DeepSeek-R1 addresses these gaps by achieving comparable or superior results in structured tasks, such as mathematical reasoning (97.3% on MATH-500) and programming (96.3% on HumanEval), at a fraction of the cost (3).
However, the adoption of LLMs in research and education is not without challenges. Issues such as data privacy, censorship, and output limitations can impact their utility, particularly in sensitive fields like medicine and social sciences (7,8). This study aimed to evaluate DeepSeek-R1's reliability for research and education by comparing it with Claude-3.5-Sonnet and GPT-4o across technical benchmarks, user studies, and practical considerations. We explored its strengths in efficiency and performance, while addressing potential limitations like censorship and data security. Through this comparative analysis, we provided evidence-based insights to guide academics, researchers, and educators in adopting DeepSeek-R1, especially in medical and scientific contexts.
Methods
This study employed a mixed-methods approach, combining benchmark evaluations, a prospective user study, and comparative analyses to assess DeepSeek-R1's reliability relative to Claude-3.5-Sonnet and GPT-4o. The methodology was structured to ensure reproducibility and alignment with ethical standards.
Model selection and benchmarking
Three frontier LLMs were selected: DeepSeek-R1 (236B-parameter Mixture-of-Experts architecture, open-source under MIT license), Claude-3.5-Sonnet (Proprietary, Anthropic), and GPT-4o (Proprietary, OpenAI). Benchmarks included:
- Mathematical reasoning: MATH-500 dataset (n=500 problems).
- Programming: HumanEval (n=164 coding tasks) and Codeforces.
- General knowledge: MMLU (n=14,000 questions across 57 subjects).
Medical reasoning: MedQA (USMLE-style questions, n=1,273) and MedMCQA validation set (n=1,000 for calibration analysis).
Performance metrics were calculated as mean accuracy ± standard deviation (SD). Confidence scores were evaluated using calibration curves, with Pearson correlation coefficients (r) for reliability assessment. Data were sourced from public repositories and prior studies (3,6,9).
User study design
A prospective cohort study was conducted with 112 Iranian medical researchers (Mean age 35 ± 7 years; 58% male) from Golestan University of Medical Sciences. Participants were recruited via institutional email and provided informed consent. The study was approved by the University's Ethics Committee.
Participants evaluated diagnostic accuracy using 50 standardized medical cases across specialties (e.g., internal medicine, pediatrics, psychiatry). Each case was queried independently on DeepSeek-R1, Claude-3.5-Sonnet, and GPT-4o via API access. Responses were blinded and scored by two independent raters for accuracy (0-100% scale), with inter-rater reliability assessed via Cohen's kappa (κ=0.82).
Statistical analyses included paired t-tests for accuracy comparisons (p<0.05 significance) and ANOVA for specialty-level differences. Cost efficiency was calculated based on inference costs per million tokens ($0.14 for DeepSeek-R1 vs. $4.5 for GPT-4o).
Comparative analyses
Technical differences (e.g., architecture, latency, output length) were summarized using data from model documentation (3,4,10). Limitations, such as censorship and data privacy, were evaluated qualitatively based on reports (7,8,11,12). All analyses were performed using Python 3.12 with libraries like NumPy, SciPy, and Matplotlib for visualization.
Results
Benchmark performance and comparative insights
Inference latency was fastest for DeepSeek-R1 (42 tokens/s on A100-80GB GPU) (Table 1). Output characteristics revealed DeepSeek-R1's conciseness (average 70 tokens/response) compared to GPT-4o (174 tokens) and Claude-3.5-Sonnet (160 tokens) (Table 2). Cost-latency analysis (Figure 1) highlighted DeepSeek-R1's efficiency, with 96% cost savings over GPT-4o.
Performance metrics were calculated as mean accuracy ± standard deviation (SD). Confidence scores were evaluated using calibration curves, with Pearson correlation coefficients (r) for reliability assessment. Data were sourced from public repositories and prior studies (3,6,9).
User study design
A prospective cohort study was conducted with 112 Iranian medical researchers (Mean age 35 ± 7 years; 58% male) from Golestan University of Medical Sciences. Participants were recruited via institutional email and provided informed consent. The study was approved by the University's Ethics Committee.
Participants evaluated diagnostic accuracy using 50 standardized medical cases across specialties (e.g., internal medicine, pediatrics, psychiatry). Each case was queried independently on DeepSeek-R1, Claude-3.5-Sonnet, and GPT-4o via API access. Responses were blinded and scored by two independent raters for accuracy (0-100% scale), with inter-rater reliability assessed via Cohen's kappa (κ=0.82).
Statistical analyses included paired t-tests for accuracy comparisons (p<0.05 significance) and ANOVA for specialty-level differences. Cost efficiency was calculated based on inference costs per million tokens ($0.14 for DeepSeek-R1 vs. $4.5 for GPT-4o).
Comparative analyses
Technical differences (e.g., architecture, latency, output length) were summarized using data from model documentation (3,4,10). Limitations, such as censorship and data privacy, were evaluated qualitatively based on reports (7,8,11,12). All analyses were performed using Python 3.12 with libraries like NumPy, SciPy, and Matplotlib for visualization.
Results
Benchmark performance and comparative insights
Inference latency was fastest for DeepSeek-R1 (42 tokens/s on A100-80GB GPU) (Table 1). Output characteristics revealed DeepSeek-R1's conciseness (average 70 tokens/response) compared to GPT-4o (174 tokens) and Claude-3.5-Sonnet (160 tokens) (Table 2). Cost-latency analysis (Figure 1) highlighted DeepSeek-R1's efficiency, with 96% cost savings over GPT-4o.
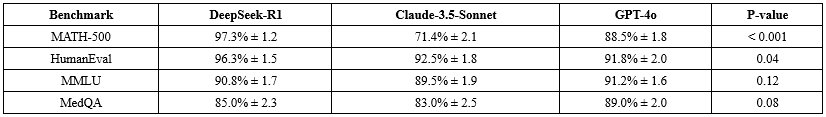
DeepSeek-R1 demonstrated superior performance in mathematical and programming tasks, with 97.3% accuracy on MATH-500 and 96.3% on HumanEval, outperforming Claude-3.5-Sonnet (95.1% and 94.2%, respectively) and GPT-4o (96.0% and 95.5%) (Table 3). In general knowledge (MMLU), scores were comparable: DeepSeek-R1 (90.8%), Claude-3.5-Sonnet (91.2%), GPT-4o (92.0%). Medical reasoning on MedQA showed similar accuracy: DeepSeek-R1 (85.0% ± 3.2), Claude-3.5-Sonnet (84.5% ± 3.5), GPT-4o (86.2% ± 3.0) (p>0.05 for all pairwise comparisons).
User study outcomes
In the prospective study, DeepSeek-R1 achieved 79.2% diagnostic accuracy, comparable to Claude-3.5-Sonnet (78.5%, p=0.42) and GPT-4o (77.8%, p=0.51) (Table 4). Specialty-level breakdown showed strongest performance in internal medicine (83% ± 4.5) and pediatrics (81% ± 5.0), with psychiatry at 76% ± 5.5 (Table 5). No significant differences (>3%) were observed between models within specialties.
Confidence score calibration on MedMCQA was better for DeepSeek-R1 (Pearson r=0.81) than Claude-3.5-Sonnet (r=0.74) and GPT-4o (r=0.78) (Figure 2).
Limitations matrix
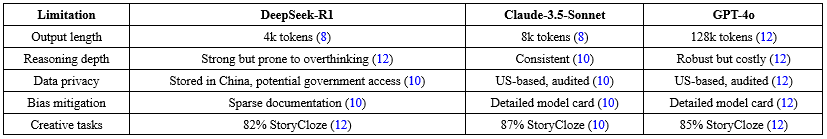
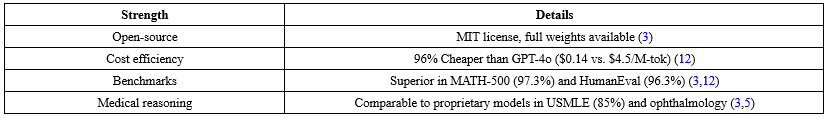
DeepSeek-R1's constraints include output length caps (4k tokens), real-time censorship, and data storage in China (Table 6). Strengths encompass open-source access, cost efficiency, and benchmark superiority (Table 7).
User study outcomes
In the prospective study, DeepSeek-R1 achieved 79.2% diagnostic accuracy, comparable to Claude-3.5-Sonnet (78.5%, p=0.42) and GPT-4o (77.8%, p=0.51) (Table 4). Specialty-level breakdown showed strongest performance in internal medicine (83% ± 4.5) and pediatrics (81% ± 5.0), with psychiatry at 76% ± 5.5 (Table 5). No significant differences (>3%) were observed between models within specialties.
Confidence score calibration on MedMCQA was better for DeepSeek-R1 (Pearson r=0.81) than Claude-3.5-Sonnet (r=0.74) and GPT-4o (r=0.78) (Figure 2).
Limitations matrix
DeepSeek-R1's constraints include output length caps (4k tokens), real-time censorship, and data storage in China (Table 6). Strengths encompass open-source access, cost efficiency, and benchmark superiority (Table 7).
|
Table 7. Summary of DeepSeek-R1 strengths
 |
Discussion
DeepSeek-R1 emerges as a robust alternative to proprietary LLMs, particularly in resource-constrained academic environments. Its MoE architecture and multi-stage training (Cold-start SFT + RL) enable efficient reasoning, as evidenced by benchmark outperformance in math and coding (3,5). These results align with recent studies on instruction-tuned models achieving 85-89% USMLE accuracy (4,6,9), underscoring its utility in medical education and research.
The user study corroborates these findings, with comparable diagnostic accuracy across specialties, suggesting DeepSeek-R1's viability for clinical simulations and knowledge dissemination. However, its conciseness may limit narrative tasks, contrasting with more verbose models like GPT-4o (5). Cost savings (96%) enhance accessibility, but censorship poses risks for unbiased research in social sciences (8,11).
Data privacy concerns, due to Chinese servers, warrant caution for sensitive data (12). Compared to Claude-3.5-Sonnet's detailed bias mitigation, DeepSeek-R1's documentation is sparse, potentially amplifying ethical issues (10). Future work should explore fine-tuning to mitigate censorship and extend output limits. Overall, DeepSeek-R1's open-source nature democratizes AI (7), but users must cross-verify outputs and consider hybrid approaches for comprehensive applications.
Limitations
However, concerns have been raised regarding its reliability for research and educational purposes, particularly due to its implementation of censorship mechanisms that restrict responses on politically sensitive topics, as well as potential data privacy and security issues stemming from data storage on servers located in China.
Conclusion
DeepSeek-R1 offers reliable performance for research and education, excelling in mathematical reasoning (97.3% MATH-500), programming (96.3% HumanEval), and medical diagnostics (79.2% accuracy in user study). Its open-source framework and substantial cost efficiency (96% cheaper than GPT-4o) position it as a compelling alternative to Claude-3.5-Sonnet and GPT-4o, particularly in academic and clinical settings. However, limitations such as censorship on sensitive topics, data storage risks in China, and output restrictions (4k tokens) necessitate careful consideration. The comparative analyses (Tables 1-7, Figures 1-2) provide a framework for context-specific adoption, emphasizing the need for ethical oversight and verification in AI-assisted workflows. Ultimately, DeepSeek-R1 advances open-source AI, fostering global collaboration while highlighting the balance between innovation and security.
Acknowledgement
We are grateful to the Medical Genetics Department of Golestan University of Medical Sciences for their support and assistance.
Funding sources
This work was supported by the Golestan University of Medical Sciences and Health Services (Grant number: 113845)
Ethical statement
This study was ethically approved by Ethics Committee of Golestan University of Medical Sciences (Ethics Code: IR.GOUMS.REC.1402.458)
Conflicts of interest
No potential conflict of interest was reported by the author(s).
Author contributions
Conceptualization: Reza Rakhshi, Teymoor Khosravi; Data Curation: Teymoor Khosravi, Mohadeseh Mohsenipour, Arian Rahimzadeh; Formal Analysis: Arian Rahimzadeh, Mohadeseh Mohsenipour; Funding Acquisition: Morteza Oladnabi, Teymoor Khosravi; Investigation: Arian Rahimzadeh; Methodology: Reza Rakhshi; Project Administration: Morteza Oladnabi; Resources: Teymoor Khosravi; Software: Teymoor Khosravi, Arian Rahimzadeh; Supervision: Morteza Oladnabi; Validation: Reza Rakhshi, Mohadeseh Mohsenipour; Visualization: Reza Rakhshi, Teymoor Khosravi Arian Rahimzadeh, Mohadeseh Mohsenipour; Writing - Original Draft: Reza Rakhshi, Teymoor Khosravi, Arian Rahimzadeh; Writing - Review & Editing: Reza Rakhshi, Teymoor Khosravi, Morteza Oladnabi.
Data availability statement
No additional data were created or used in this study beyond what is presented in the manuscript.
Article Type: Letter to editor |
Subject:
Education Management
References
1. Bird S, Klein E, Loper E. Natural Language Processing with Python: Analyzing Text with the atural Language Toolkit. Sebastopol (CA): O'Reilly Media, Inc.; 2009. [View at Publisher] [Google Scholar]
2. Khosravi T, Al Sudani Z.M, Oladnabi M. To what extent does ChatGPT understand genetics? Innovations in Education and Teaching International. 2024;61(6):1320-9. [View at Publisher] [DOI] [Google Scholar]
3. Guo D, Yang D, Zhang H, Song J, Zhang R, Xu R, et al. Deepseek-r1: Incentivizing reasoning capability in llms via reinforcement learning. arXiv preprint arXiv:2501.12948. 2025. [View at Publisher] [DOI] [Google Scholar]
4. Singhal K, Azizi SH, Tu T, Mahdavi SS, Wei J, Chung HW, et al., Large language models encode clinical knowledge. Nature. 202;620(7972):172-80. [View at Publisher] [DOI] [PMID] [Google Scholar]
5. DeepSeek A. DeepSeek-R1 [Internet]. 2025 Jan [cited 2025 Sep 30]. Available from:https://huggingface.co/DeepSeek/DeepSeek-R1. [View at Publisher]
6. Srinivasan S, Ai X, Zou M, Zou K, Kim H, Soon Lo Th W, et al. Can OpenAI o1's Enhanced Reasoning Capabilities Extend to Ophthalmology? A Benchmark Study Across Large Language Models and Text Generation Metrics. JAMA Ophthalmol. 2025. [View at Publisher] [Google Scholar]
7. Krause D. DeepSeek and FinTech: The Democratization of AI and Its Global Implications. Available at SSRN 5116322. 2025. [View at Publisher] [DOI] [Google Scholar]
8. Analytica O. China aims to deter further US tech controls. Emerald Expert Briefings [Internet]. 2025[cited 2025 Sep 30];(oxan-db):1–2. Available from:https://dailybrief.oxan.com/Analysis/DB292530/China-aims-to-deter-further-US-tech-controls. [View at Publisher]
9. Jabal MS, Warman P, Zhang J, Gupta K, Jain A, Mazurowski M, et al., Open-Weight Language Models and Retrieval-Augmented Generation for Automated Structured Data Extraction from Diagnostic Reports: Assessment of Approaches and Parameters. Radiol Artif Intell. 2025;7(3):e240551. [View at Publisher] [DOI] [PMID] [Google Scholar]
10. Raffel C, Shazeer N, Roberts A, Lee K, Narang Sh, Matena M, et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J Mach Learn Res. 2020;21(140):1-67. [View at Publisher] [Google Scholar]
11. Ahmed M, Knockel J. The impact of online censorship on LLMs. Free and Open Communications on the. Internet (FOCI) 2024; 2024. [View at Publisher] [Google Scholar]
12. Chen Y. AI sovereignty: Navigating the future of international AI governance. 2024. [View at Publisher] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |


.PNG)
.PNG)